1. Introduction
The desired input of many comparative studies is a matrix that includes all the data available for a set of traits and a focal taxon. Bringing similar types of trait data together across studies is notoriously difficult, but is straightforward with ontology annotations. Trait data that are tagged using similar ontology terms can be automatically called using R functions.
This workshop uses trait data for vertebrates that has been annotated (tagged) with ontology terms. These data are accessed from the Phenoscape KB. In this lesson we use R functions to obtain and view character matrices from the KB, obtain synthetic matrices of inferred presence/absence characters, and understand the meaning and relations of ontology terms. The lesson assumes basic knowledge of R and RStudio.
2. Browse and filter data in the KB to understand the scope of available data.
You would like to understand the evolution of morphological traits in a taxonomic group, and you need a matrix to map to your (typically molecular) phylogenetic tree. You come to the Phenoscape KB to see what types of data are available across the members of your clade. In this exercise, you are an ichthyologist, interested in catfishes (Siluriformes), specifically the bullhead catfishes in the family Ictaluridae.
You begin by querying ‘Siluriformes’ in the Phenoscape KB faceted browsing page. You immediately see the scope of the data:
From left to right across the top tabs, for ‘Siluriformes’:
- There are 4,969 unique phenotypes
- There are 292,853 taxon annotations
- There are 1,505 siluriform taxa with data in the KB
- There are 40 publications in the KB that include catfish phenotypes
- There are 19,232 candidate genes linked to the phenotypes
Taxon annotations are the ontology terms tagged to original free text descriptions of phenotypes for each siluriform taxon. Click on the ‘Taxon annotations’ tab, and then the ‘Sources’ box on the right hand side of each row to see the publication source(s) and free text description of the phenotype to which each ontology annotation was tagged.
You are specifically interested in Ictaluridae (bullhead catfishes), and you click on that family in the faceted browsing interface. Now filter using ‘fin’ as the anatomical entity under ‘Query’ and include parts (check the ‘parts’ box.)
Download these taxa and their morphological trait data using the link provided.
Go to the publications tab, which shows nine publications that contain fin data. Note that any of these studies, e.g., Lundberg (1992), can be entered under ‘publications’ in faceted browsing, but the original matrix cannot be downloaded.
3. Retrieve term info using RPhenoscape
Now we will repeat some of the above steps in R, beginning with a query for ‘Ictaluridae’ and ‘fin’ (including parts) to retrieve the list of studies that contain fin characters for this taxon.
The RPhenoscape package provides convenient access to the Phenoscape Knowledgebase in the form of R functions returning R-native data structures. So that we don’t have to prefix every call of the package’s functions with the package name, we will first attach the package:
First let’s look up information about the query terms. The find_term()
function searches the KB for terms matching a given search text, and
will return the ID (term IRI), label, source ontology IRI, and type of
match (e.g., exact, broad). For example, search for terms containing the
string “fin” in the label or synonym:
find_term("fin")
#> id
#> 1 http://purl.obolibrary.org/obo/UBERON_0008897
#> 2 http://purl.obolibrary.org/obo/UBERON_0002389
#> 3 http://purl.obolibrary.org/obo/UBERON_0009565
#> 4 http://purl.obolibrary.org/obo/UBERON_0009552
#> 5 http://purl.obolibrary.org/obo/ZFA_0000108
#> 6 http://purl.obolibrary.org/obo/UBERON_2005316
#> 7 http://purl.obolibrary.org/obo/GO_0033333
#> 8 http://purl.obolibrary.org/obo/GO_0033334
#> 9 http://purl.obolibrary.org/obo/GO_0031101
#> 10 http://purl.obolibrary.org/obo/HP_0001231
#> 11 http://purl.obolibrary.org/obo/HP_0001161
#> 12 http://purl.obolibrary.org/obo/HP_0001187
#> 13 http://purl.obolibrary.org/obo/HP_0001238
#> 14 http://purl.obolibrary.org/obo/HP_0001166
#> 15 http://purl.obolibrary.org/obo/HP_0001182
#> 16 http://purl.obolibrary.org/obo/HP_0001817
#> 17 http://purl.obolibrary.org/obo/HP_0001500
#> 18 http://purl.obolibrary.org/obo/HP_0001821
#> 19 http://purl.obolibrary.org/obo/HP_0004095
#> 20 http://purl.obolibrary.org/obo/HP_0004097
#> 21 http://purl.obolibrary.org/obo/HP_0002213
#> 22 http://purl.obolibrary.org/obo/HP_0004099
#> 23 http://purl.obolibrary.org/obo/HP_0006101
#> 24 http://purl.obolibrary.org/obo/HP_0006107
#> 25 http://purl.obolibrary.org/obo/HP_0001812
#> 26 http://purl.obolibrary.org/obo/HP_0001804
#> 27 http://purl.obolibrary.org/obo/HP_0005867
#> 28 http://purl.obolibrary.org/obo/HP_0007477
#> 29 http://purl.obolibrary.org/obo/HP_0009380
#> 30 http://purl.obolibrary.org/obo/HP_0008391
#> 31 http://purl.obolibrary.org/obo/HP_0010525
#> 32 http://purl.obolibrary.org/obo/HP_0009700
#> 33 http://purl.obolibrary.org/obo/HP_0009381
#> 34 http://purl.obolibrary.org/obo/HP_0010557
#> 35 http://purl.obolibrary.org/obo/HP_0008402
#> 36 http://purl.obolibrary.org/obo/HP_0010554
#> 37 http://purl.obolibrary.org/obo/HP_0025131
#> 38 http://purl.obolibrary.org/obo/HP_0025261
#> 39 http://purl.obolibrary.org/obo/HP_0011300
#> 40 http://purl.obolibrary.org/obo/HP_0031090
#> 41 http://purl.obolibrary.org/obo/HP_0030367
#> 42 http://purl.obolibrary.org/obo/HP_0030837
#> 43 http://purl.obolibrary.org/obo/HP_0030771
#> 44 http://purl.obolibrary.org/obo/HP_0030033
#> 45 http://purl.obolibrary.org/obo/HP_0030029
#> 46 http://purl.obolibrary.org/obo/HP_0012742
#> 47 http://purl.obolibrary.org/obo/HP_0012276
#> 48 http://purl.obolibrary.org/obo/HP_0100759
#> 49 http://purl.obolibrary.org/obo/HP_0034037
#> 50 http://purl.obolibrary.org/obo/HP_0045089
#> 51 http://purl.obolibrary.org/obo/HP_0100798
#> 52 http://purl.obolibrary.org/obo/HP_0040019
#> 53 http://purl.obolibrary.org/obo/HP_0100807
#> 54 http://purl.obolibrary.org/obo/HP_0045090
#> 55 http://purl.obolibrary.org/obo/HP_0033976
#> 56 http://purl.obolibrary.org/obo/MP_0000561
#> 57 http://purl.obolibrary.org/obo/MP_0000562
#> 58 http://purl.obolibrary.org/obo/MP_0003807
#> 59 http://purl.obolibrary.org/obo/MP_0000564
#> 60 http://purl.obolibrary.org/obo/MP_0002544
#> 61 http://purl.obolibrary.org/obo/MP_0006296
#> 62 http://purl.obolibrary.org/obo/MP_0009930
#> 63 http://purl.obolibrary.org/obo/MP_0030929
#> 64 http://purl.obolibrary.org/obo/MP_0030930
#> 65 http://purl.obolibrary.org/obo/UBERON_4000172
#> 66 http://purl.obolibrary.org/obo/UBERON_4400005
#> 67 http://purl.obolibrary.org/obo/ZFA_0001058
#> 68 http://purl.obolibrary.org/obo/VTO_0013147
#> 69 http://purl.obolibrary.org/obo/VTO_0003107
#> 70 http://purl.obolibrary.org/obo/VTO_0021975
#> 71 http://purl.obolibrary.org/obo/VTO_0018051
#> 72 http://purl.obolibrary.org/obo/VTO_0016713
#> 73 http://purl.obolibrary.org/obo/VTO_0015894
#> 74 http://purl.obolibrary.org/obo/VTO_0023422
#> 75 http://purl.obolibrary.org/obo/VTO_0023456
#> 76 http://purl.obolibrary.org/obo/VTO_0028227
#> 77 http://purl.obolibrary.org/obo/VTO_0023261
#> 78 http://purl.obolibrary.org/obo/VTO_0025094
#> 79 http://purl.obolibrary.org/obo/VTO_0023240
#> 80 http://purl.obolibrary.org/obo/VTO_0024763
#> 81 http://purl.obolibrary.org/obo/VTO_0029180
#> 82 http://purl.obolibrary.org/obo/VTO_0031838
#> 83 http://purl.obolibrary.org/obo/VTO_0059371
#> 84 http://purl.obolibrary.org/obo/VTO_0059142
#> 85 http://purl.obolibrary.org/obo/VTO_0063879
#> 86 http://purl.obolibrary.org/obo/VTO_0062623
#> 87 http://purl.obolibrary.org/obo/VTO_0073037
#> 88 http://purl.obolibrary.org/obo/VTO_0066755
#> 89 http://purl.obolibrary.org/obo/VTO_0069689
#> 90 http://purl.obolibrary.org/obo/VTO_0068161
#> 91 http://purl.obolibrary.org/obo/VTO_0073687
#> label
#> 1 fin
#> 2 manual digit
#> 3 nail of manual digit
#> 4 distal segment of manual digit
#> 5 fin (zebrafish)
#> 6 fin fold pectoral fin bud
#> 7 fin development
#> 8 fin morphogenesis
#> 9 fin regeneration
#> 10 Abnormal fingernail morphology
#> 11 Hand polydactyly
#> 12 Hyperextensibility of the finger joints
#> 13 Slender finger
#> 14 Arachnodactyly
#> 15 Tapered finger
#> 16 Absent fingernail
#> 17 Broad finger
#> 18 Broad nail
#> 19 Curved fingers
#> 20 Deviation of finger
#> 21 Fine hair
#> 22 Macrodactyly
#> 23 Finger syndactyly
#> 24 Fingerpad telangiectases
#> 25 Hyperconvex fingernails
#> 26 Hypoplastic fingernail
#> 27 4-5 metacarpal synostosis
#> 28 Abnormal dermatoglyphics
#> 29 Aplasia of the fingers
#> 30 Dystrophic fingernails
#> 31 Finger agnosia
#> 32 Finger symphalangism
#> 33 Short finger
#> 34 Overlapping fingers
#> 35 Ridged fingernail
#> 36 Cutaneous finger syndactyly
#> 37 Finger swelling
#> 38 Stiff finger
#> 39 Broad fingertip
#> 40 Finger dactylitis
#> 41 Finger hyperphalangy
#> 42 Finger pain
#> 43 Mallet finger
#> 44 Small finger
#> 45 Splayed fingers
#> 46 Thin fingernail
#> 47 Digital flexor tenosynovitis
#> 48 Clubbing of fingers
#> 49 Pseudo-chilblains on fingers
#> 50 Distinctive finding
#> 51 Fingernail dysplasia
#> 52 Finger clinodactyly
#> 53 Long fingers
#> 54 Minor finding
#> 55 Volar fingernail
#> 56 adactyly
#> 57 polydactyly
#> 58 camptodactyly
#> 59 syndactyly
#> 60 brachydactyly
#> 61 arachnodactyly
#> 62 fuzzy hair
#> 63 increased digit pigmentation
#> 64 decreased digit pigmentation
#> 65 lepidotrichium
#> 66 fin ray
#> 67 caudal fin (zebrafish)
#> 68 Callosciurus finlaysonii
#> 69 Limnonectes finchi
#> 70 Agama finchi
#> 71 Plestiodon finitimus
#> 72 Uroplatus finiavana
#> 73 Varanus finschi
#> 74 Amazona finschi
#> 75 Aratinga finschi
#> 76 Euphonia finschi
#> 77 Micropsitta finschii
#> 78 Oenanthe finschii
#> 79 Psittacula finschii
#> 80 Pycnonotus finlaysoni
#> 81 Stizorhina finschi
#> 82 Francolinus finschi
#> 83 Bunocephalus sp. (Fink and Fink 1981)
#> 84 Cnemidocarpa finmarkiensis
#> 85 Garra findolabium
#> 86 Schistura finis
#> 87 Lamprologus finalimus
#> 88 Saurenchelys finitimus
#> 89 Soleonasus finis
#> 90 Theragra finnmarchica
#> 91 Benitochromis finleyi
#> isDefinedBy matchType
#> 1 http://purl.obolibrary.org/obo/uberon.owl exact
#> 2 http://purl.obolibrary.org/obo/uberon.owl broad
#> 3 http://purl.obolibrary.org/obo/uberon.owl broad
#> 4 http://purl.obolibrary.org/obo/uberon.owl broad
#> 5 http://purl.obolibrary.org/obo/zfa.owl partial
#> 6 http://purl.obolibrary.org/obo/uberon.owl partial
#> 7 http://purl.obolibrary.org/obo/go.owl partial
#> 8 http://purl.obolibrary.org/obo/go.owl partial
#> 9 http://purl.obolibrary.org/obo/go.owl partial
#> 10 http://purl.obolibrary.org/obo/hp.owl partial
#> 11 http://purl.obolibrary.org/obo/hp.owl broad
#> 12 http://purl.obolibrary.org/obo/hp.owl partial
#> 13 http://purl.obolibrary.org/obo/hp.owl partial
#> 14 http://purl.obolibrary.org/obo/hp.owl broad
#> 15 http://purl.obolibrary.org/obo/hp.owl partial
#> 16 http://purl.obolibrary.org/obo/hp.owl partial
#> 17 http://purl.obolibrary.org/obo/hp.owl partial
#> 18 http://purl.obolibrary.org/obo/hp.owl broad
#> 19 http://purl.obolibrary.org/obo/hp.owl partial
#> 20 http://purl.obolibrary.org/obo/hp.owl partial
#> 21 http://purl.obolibrary.org/obo/hp.owl partial
#> 22 http://purl.obolibrary.org/obo/hp.owl broad
#> 23 http://purl.obolibrary.org/obo/hp.owl partial
#> 24 http://purl.obolibrary.org/obo/hp.owl partial
#> 25 http://purl.obolibrary.org/obo/hp.owl partial
#> 26 http://purl.obolibrary.org/obo/hp.owl partial
#> 27 http://purl.obolibrary.org/obo/hp.owl broad
#> 28 http://purl.obolibrary.org/obo/hp.owl broad
#> 29 http://purl.obolibrary.org/obo/hp.owl partial
#> 30 http://purl.obolibrary.org/obo/hp.owl partial
#> 31 http://purl.obolibrary.org/obo/hp.owl partial
#> 32 http://purl.obolibrary.org/obo/hp.owl partial
#> 33 http://purl.obolibrary.org/obo/hp.owl partial
#> 34 http://purl.obolibrary.org/obo/hp.owl partial
#> 35 http://purl.obolibrary.org/obo/hp.owl partial
#> 36 http://purl.obolibrary.org/obo/hp.owl partial
#> 37 http://purl.obolibrary.org/obo/hp.owl partial
#> 38 http://purl.obolibrary.org/obo/hp.owl partial
#> 39 http://purl.obolibrary.org/obo/hp.owl partial
#> 40 http://purl.obolibrary.org/obo/hp.owl partial
#> 41 http://purl.obolibrary.org/obo/hp.owl partial
#> 42 http://purl.obolibrary.org/obo/hp.owl partial
#> 43 http://purl.obolibrary.org/obo/hp.owl partial
#> 44 http://purl.obolibrary.org/obo/hp.owl partial
#> 45 http://purl.obolibrary.org/obo/hp.owl partial
#> 46 http://purl.obolibrary.org/obo/hp.owl partial
#> 47 http://purl.obolibrary.org/obo/hp.owl broad
#> 48 http://purl.obolibrary.org/obo/hp.owl partial
#> 49 http://purl.obolibrary.org/obo/hp.owl partial
#> 50 http://purl.obolibrary.org/obo/hp.owl partial
#> 51 http://purl.obolibrary.org/obo/hp.owl partial
#> 52 http://purl.obolibrary.org/obo/hp.owl partial
#> 53 http://purl.obolibrary.org/obo/hp.owl partial
#> 54 http://purl.obolibrary.org/obo/hp.owl partial
#> 55 http://purl.obolibrary.org/obo/hp.owl partial
#> 56 http://purl.obolibrary.org/obo/mp.owl broad
#> 57 http://purl.obolibrary.org/obo/mp.owl broad
#> 58 http://purl.obolibrary.org/obo/mp.owl broad
#> 59 http://purl.obolibrary.org/obo/mp.owl broad
#> 60 http://purl.obolibrary.org/obo/mp.owl broad
#> 61 http://purl.obolibrary.org/obo/mp.owl broad
#> 62 http://purl.obolibrary.org/obo/mp.owl broad
#> 63 http://purl.obolibrary.org/obo/mp.owl broad
#> 64 http://purl.obolibrary.org/obo/mp.owl broad
#> 65 http://purl.obolibrary.org/obo/uberon.owl broad
#> 66 http://purl.obolibrary.org/obo/uberon.owl partial
#> 67 http://purl.obolibrary.org/obo/zfa.owl partial
#> 68 http://purl.obolibrary.org/obo/vto.owl partial
#> 69 http://purl.obolibrary.org/obo/vto.owl partial
#> 70 http://purl.obolibrary.org/obo/vto.owl partial
#> 71 http://purl.obolibrary.org/obo/vto.owl partial
#> 72 http://purl.obolibrary.org/obo/vto.owl partial
#> 73 http://purl.obolibrary.org/obo/vto.owl partial
#> 74 http://purl.obolibrary.org/obo/vto.owl partial
#> 75 http://purl.obolibrary.org/obo/vto.owl partial
#> 76 http://purl.obolibrary.org/obo/vto.owl partial
#> 77 http://purl.obolibrary.org/obo/vto.owl partial
#> 78 http://purl.obolibrary.org/obo/vto.owl partial
#> 79 http://purl.obolibrary.org/obo/vto.owl partial
#> 80 http://purl.obolibrary.org/obo/vto.owl partial
#> 81 http://purl.obolibrary.org/obo/vto.owl partial
#> 82 http://purl.obolibrary.org/obo/vto.owl partial
#> 83 http://purl.obolibrary.org/obo/vto.owl partial
#> 84 http://purl.obolibrary.org/obo/vto.owl partial
#> 85 http://purl.obolibrary.org/obo/vto.owl partial
#> 86 http://purl.obolibrary.org/obo/vto.owl partial
#> 87 http://purl.obolibrary.org/obo/vto.owl partial
#> 88 http://purl.obolibrary.org/obo/vto.owl partial
#> 89 http://purl.obolibrary.org/obo/vto.owl partial
#> 90 http://purl.obolibrary.org/obo/vto.owl partial
#> 91 http://purl.obolibrary.org/obo/vto.owl partialNote that because the KB integrates model organism phenotypes, the results show types of fin specific to model organisms, such as “fin (zebrafish)”.
There are several parameters available to narrow and filter search
results. For example, the parameter matchType allows
restricting to exact matches only:
find_term("fin", matchTypes = c("exact"))
#> id label
#> 1 http://purl.obolibrary.org/obo/UBERON_0008897 fin
#> isDefinedBy matchType
#> 1 http://purl.obolibrary.org/obo/uberon.owl exactTo retrieve more details about a given term, such as definition,
synonyms and classification, you can pass the term IRI (e.g., ‘http://purl.obolibrary.org/obo/UBERON_0008897’ for fin)
to the function as.terminfo().
Classification details include the superclass (parent) and subclass
(child) relationships for a given ontology term, along with other
relationships such as part_of.
For example, view the relationships and term info for “pectoral fin”:
pecfin <- find_term('pectoral fin', matchType='exact')
as.terminfo(pecfin)
#> terminfo 'pectoral fin' http://purl.obolibrary.org/obo/UBERON_0000151
#> Definition: Paired fin that is located in the thoracic region of the body.
#> Synonyms:
#> forefin (exact)
#> pectoral fins (related)
#> Relationships:
#> has skeleton pectoral fin skeleton
#> develops from pectoral appendage bud
#> Subclass of:
#> paired fin
#> pectoral appendage
#> Superclass of:
#> archipterygial fin
#> pectoral fin (zebrafish)
#> pectoral fin and (part_of some larva)
#> pectoral fin and (part_of some larva)
#> pectoral fin and (part_of some larval stage)
#> pectoral fin and (part_of some larval stage)For a specific taxon, such as Siluriformes, you can also view the taxon rank, whether or not the taxon is extinct, and its common name:
taxon <- find_term('Siluriformes')
as.terminfo(taxon)
#> terminfo 'Siluriformes' http://purl.obolibrary.org/obo/VTO_0034991
#> Synonyms:
#> catfish (related)
#> catfishes (related)
#> Subclass of:
#> Otophysi
#> Superclass of:
#> Ailia
#> Akysidae
#> Amblycipitidae
#> Amphiliidae
#> Anchariidae
#> Ariidae
#> Aspredinidae
#> Astroblepidae
#> Auchenipteridae
#> Auchenoglanididae
#> Australoglanididae
#> Austroglanididae
#> Bagridae
#> Callichthyidae
#> Cetopsidae
#> Chacidae
#> Clariidae
#> Claroteidae
#> Clupisoma
#> Conorhynchidae
#> Cranoglanididae
#> Diplomystidae
#> Doradidae
#> Eopeyeria
#> Erethistidae
#> Eutropiichthys
#> Heptapteridae
#> Heteropneustidae
#> Horabagridae
#> Hypophthalmidae
#> Hypsidoridae
#> Ictaluridae
#> Lacantuniidae
#> Laides
#> Liobagrus
#> Loricariidae
#> Malapteruridae
#> Mochokidae
#> Nematogenyidae
#> Neotropius
#> Olyridae
#> Pangasiidae
#> Pareutropius
#> Phreatobius
#> Pimelodidae
#> Platytropius
#> Plotosidae
#> Proeutropiichthys
#> Pseudopimelodidae
#> Rita
#> Schilbidae
#> Scoloplacidae
#> Selenaspis
#> Silonia
#> Siluridae
#> Siluroidei
#> Sisoridae
#> Trichomycteridae
#> Xiurenbagrus
#> Extinct: FALSE
#> Rank: order
#> Common Name: catfish4. Retrieve study matrices using RPhenoscape
Now we will query for ‘Ictaluridae’ and ‘fin’ (including parts) to retrieve the list of studies that contain fin characters for this taxon.
slist <- get_studies(taxon = "Ictaluridae", entity = "fin", includeRels = "part of")The list is returned as a data.frame containing the study IDs and citations. Navigate to Global Environment and click on ‘slist’ to view the ID and citation (author, year) for each study in the KB that contains any ictalurid species associated with fin data. Note that taxon, entity, and quality are optional arguments in this query, and we have also included fin parts. Here we found nine studies. Click on ‘slist’ in the Global Environment to view these studies and their IDs.
Next we want to obtain the character matrix for one of these studies. Using the study IDs from the previous step, we can get a list of the character matrices as NeXML objects. We will choose the Lundberg (1992) publication, the fifth item on the list above.
lundberg_nexml <- get_study_data(slist$id[5])Now go to Global Environment pane and click on ‘lundberg_nexml’ to view the study ID and associated NeXML object.
To view the full Lundberg (1992) matrix within RStudio, we need to first retrieve the matrix as a data.frame from the NeXML object:
lundberg_matrix <- get_char_matrix(lundberg_nexml[[1]], otus_id = FALSE)You can take a look at a small part of the Lundberg (1992) matrix (e.g., the first five taxa and first five characters):
lundberg_matrix[1:5, 1:7]
#> taxa otu
#> 1 Ameiurus brunneus otu_331eacec-f62a-489b-aa97-01f88c469d4a
#> 2 Ameiurus catus otu_f0b4abb4-c4ca-4d99-b6d3-d069176eea82
#> 3 Ameiurus melas otu_5a3062f1-9803-4120-ae34-d2aa1ce02a75
#> 4 Ameiurus natalis otu_b92dfa0d-8c16-4228-815f-c418c6e92781
#> 5 Ameiurus nebulosus otu_31f42c8a-cd70-4bf4-994d-9bc483822ba7
#> Anal-fin rays, species mean count Anterior dentations of pectoral spine
#> 1 1 3
#> 2 2 2
#> 3 2 1
#> 4 3 1
#> 5 2 2
#> Anterior distal serrae of pectoral spine Anterior extent of sphenotic
#> 1 1 0
#> 2 2 0
#> 3 0 0
#> 4 2 0
#> 5 1 0
#> Anterior limb of fourth transverse process
#> 1 2
#> 2 1
#> 3 1
#> 4 0
#> 5 0Now navigate to Global Environment and click on ‘lundberg_matrix’ to view the matrix, including the total number of taxa and characters (i.e., the dimensions of the data.frame). To view the entire matrix in this panel, click on the first icon that appears to the right when you hover over the second row.
We can use the information in the NeXML object to convert character state symbols to labels:
state_symbols2labels(lundberg_nexml[[1]], charmat = lundberg_matrix)[1:5, 1:7]
#> taxa otu
#> 1 Ameiurus brunneus otu_331eacec-f62a-489b-aa97-01f88c469d4a
#> 2 Ameiurus catus otu_f0b4abb4-c4ca-4d99-b6d3-d069176eea82
#> 3 Ameiurus melas otu_5a3062f1-9803-4120-ae34-d2aa1ce02a75
#> 4 Ameiurus natalis otu_b92dfa0d-8c16-4228-815f-c418c6e92781
#> 5 Ameiurus nebulosus otu_31f42c8a-cd70-4bf4-994d-9bc483822ba7
#> Anal-fin rays, species mean count Anterior dentations of pectoral spine
#> 1 16.1-20 large
#> 2 20.1-25.5 moderate
#> 3 20.1-25.5 small
#> 4 25.6-28 small
#> 5 20.1-25.5 moderate
#> Anterior distal serrae of pectoral spine
#> 1 <3 moderately sharp serrae
#> 2 3-6 moderately sharp serrae
#> 3 absent or scarcely developed
#> 4 3-6 moderately sharp serrae
#> 5 <3 moderately sharp serrae
#> Anterior extent of sphenotic
#> 1 reaches or extends anterior to the level of anterior margin of epiphyseal bar
#> 2 reaches or extends anterior to the level of anterior margin of epiphyseal bar
#> 3 reaches or extends anterior to the level of anterior margin of epiphyseal bar
#> 4 reaches or extends anterior to the level of anterior margin of epiphyseal bar
#> 5 reaches or extends anterior to the level of anterior margin of epiphyseal bar
#> Anterior limb of fourth transverse process
#> 1 supracleithral facet shallow, basal recess shallow and broad
#> 2 supracleithral facet deep, basal recess deep and broad
#> 3 supracleithral facet deep, basal recess deep and broad
#> 4 supracleithral facet deep, basal recess deep and narrow
#> 5 supracleithral facet deep, basal recess deep and narrowNote that all taxa and characters from this study are returned, not just those pertaining to our original search terms. In this example, many anatomical entities other than ‘fin’ and its parts are returned as part of the original study matrix. In a later step, we will learn how to subset a matrix using anatomy or taxonomy terms.
5. Get a presence/absence character matrix using OntoTrace in RPhenoscape
In the previous step, we queried the KB for individual studies that contain characters pertaining to the fins in ictalurid catfish, and we obtained character matrices for source publications that included those characters. Now we want to get a matrix that synthesizes these characters into a single matrix.
OntoTrace from the KB combines characters from multiple publications into a single synthetic matrix. It allows a user to specify any number or combination of anatomical entities and taxa. It returns a matrix as a nexml object that includes only presence or absence of anatomical entities. It uses asserted data of presence or absence, i.e., originally described by an author, or inferred data. Data are inferred from asserted characters, e.g., if an author describes a part of a fin such as ‘fin ray’, the fin is inferred present and scored as such in the matrix.
The OntoTrace default settings are to include parts of the anatomical entity and to include variable-only characters, but here we will include those that are invariant as well.
nex <- get_ontotrace_data(taxon = "Ictaluridae", entity = "fin", variable_only = FALSE)To view the matrix within RStudio, get the matrix (m) as a
data.frame from the NeXML object. Opening this (‘m’) in the
Global Environment shows the taxa, character names, and state
assignments (0, absent; 1, present) (note: the otu and otus are
NeXML-internal IDs):
m <- get_char_matrix(nex)The character and taxon lists for the matrix can also be returned. Click on ‘meta’ in the Global Environment pane to view the data.frames for taxa and entities:
meta <- get_char_matrix_meta(nex)The above returns 29 species of ictalurid catfishes and 124 anatomical entities for fins and their parts.
You will need to go to the Phenoscape KB to view the supporting states, i.e., the source data upon which inferences were made or those that were directly asserted.
For example, in the returned OntoTrace matrix (see ‘m’ in Global Environment (row 10)) the pelvic fin is scored as present (‘1’) for Ictalurus australis. Enter these data on the faceted browsing page:
- Anatomical entity = ‘pelvic fin’
- Quality = ‘present’
- Taxon = ‘Ictalurus australis’
Looking at ‘Taxon annotations’, you will find that there are zero phenotypes, i.e., no asserted character states to ‘pelvic fin present’. Now click the ‘inferred presence’ radio button under Phenotypic quality. You will now see two phenotypes for this taxon in the KB. These phenotypes ‘pelvic fin lepidotrichium amount’ and ‘pelvic splint present’ logically imply the presence of a pelvic fin. Thus, though an author did not directly assert the presence of a pelvic fin in Ictalurus australis, it is inferred and thus reported as present (‘1’) in the OntoTrace matrix.
6. Subset the matrix taxonomically
In the above step you retrieved a synthetic matrix for the Ictaluridae. Now you would like to pare it down in taxonomic and anatomical scope. Let’s begin by trimming the matrix down to the presence and absence characters pertaining only to the genus Ictalurus. We will reduce the matrix to include only descendents of Ictalurus.
First, we need to parse the list to determine which taxa in the matrix (‘m’) are descendants of Ictalurus. This returns a list of TRUE or FALSE assignments for each taxon in the matrix:
is_desc <- is_descendant('Ictalurus', m$taxa)We then subset the matrix m to the descendants of Ictalurus:
ictalurus_m <- m[is_desc, ]View the new matrix by clicking on the ‘ictalurus_m’
data.frame in the Global Environment.
We can also subset the matrix by anatomy. In this case, let’s select
characters for the pectoral fin and its parts. We will use is_descendant()
to first parse the characters (column names; note the first three
taxon-related columns are excluded) to determine which are descendents
(types or parts) of the term ‘pectoral fin’. Because this function only
looks for descendent terms, we also need to specify that any characters
pertaining to ‘pectoral fin’ itself are included.
is_desc_pecfin <- is_descendant('pectoral fin', colnames(ictalurus_m)[-c(1,2,3)], includeRels = "part_of") | colnames(ictalurus_m)[-c(1,2,3)] == 'pectoral fin'
table(is_desc_pecfin)
#> is_desc_pecfin
#> FALSE TRUE
#> 110 14Using the ‘colnames’ command below, view the character names to verify that pectoral fin related characters will be selected:
colnames(ictalurus_m)[-c(1,2,3)][is_desc_pecfin]
#> [1] "anterior dentation of pectoral fin spine"
#> [2] "anterior distal serration of pectoral fin spine"
#> [3] "pectoral fin"
#> [4] "pectoral fin lepidotrichium"
#> [5] "pectoral fin proximal radial bone"
#> [6] "pectoral fin proximal radial cartilage"
#> [7] "pectoral fin proximal radial element"
#> [8] "pectoral fin radial bone"
#> [9] "pectoral fin radial cartilage"
#> [10] "pectoral fin radial element"
#> [11] "pectoral fin radial skeleton"
#> [12] "pectoral fin skeleton"
#> [13] "pectoral fin spine"
#> [14] "posterior dentation of pectoral fin spine"We now subset the matrix m using ‘is_desc_pecfin’ and retain the first three taxon-related columns:
to_keep <- c(TRUE, TRUE, TRUE, is_desc_pecfin)
pecfin_m <- ictalurus_m[, to_keep]View this matrix of pectoral fin characters for Ictalurus by clicking on ‘pecfin_m’ in the Global Environment.
6. Retrieve matrix of anatomical dependencies
Knowing how the information about an anatomical structure may rely on other layers of anatomical information is critical, if the goal is to isolate and analyze independent features. We can query the KB for a presence-absence dependency matrix for a specified set of anatomical structures. The dependency matrix is derived from the formalized knowledge of relationships among entities encoded in the ontology. In the example below, the presence of ‘pelvic splint’ is dependent on the presence of ‘pelvic fin’. In other words, the pelvic fin is implied present when the pelvic splint is present because ‘pelvic splint’ is part_of ‘pelvic fin’.
tl <- c("pectoral fin", "pelvic fin", "pelvic splint", "antorbital", "preopercle")
dep_mat <- pa_dep_matrix(tl, .names = "label")
dep_mat
#> pectoral fin pelvic fin pelvic splint antorbital preopercle
#> pectoral fin 1 0 0 0 0
#> pelvic fin 0 1 0 0 0
#> pelvic splint 0 1 1 0 0
#> antorbital 0 0 0 1 0
#> preopercle 0 0 0 0 1In the PARAMO tutorial, we will learn how to use these dependency matrices to reconstruct individual traits and entire phenotypes.
7. Retrieve semantic similarity matrix
We can also obtain similarity metrics for phenotypes based on ontological distance. Three similarity metrics - Jaccard, Tanimoto, and Cosine - are computed by the KB to measure similarity between terms (see RPhenoscape documentation in the Help panel for how these are measured). For example, here we compute and visualize Jaccard similarity for premaxillary tooth phenotypes in Ictaluridae:
phens <- get_phenotypes(entity = "premaxillary tooth", taxon = "Ictaluridae")
nrow(phens)
#> [1] 9
sm <- jaccard_similarity(terms = phens$id, .labels = phens$label, .colnames = "label")
plot(hclust(as.dist(1-sm)))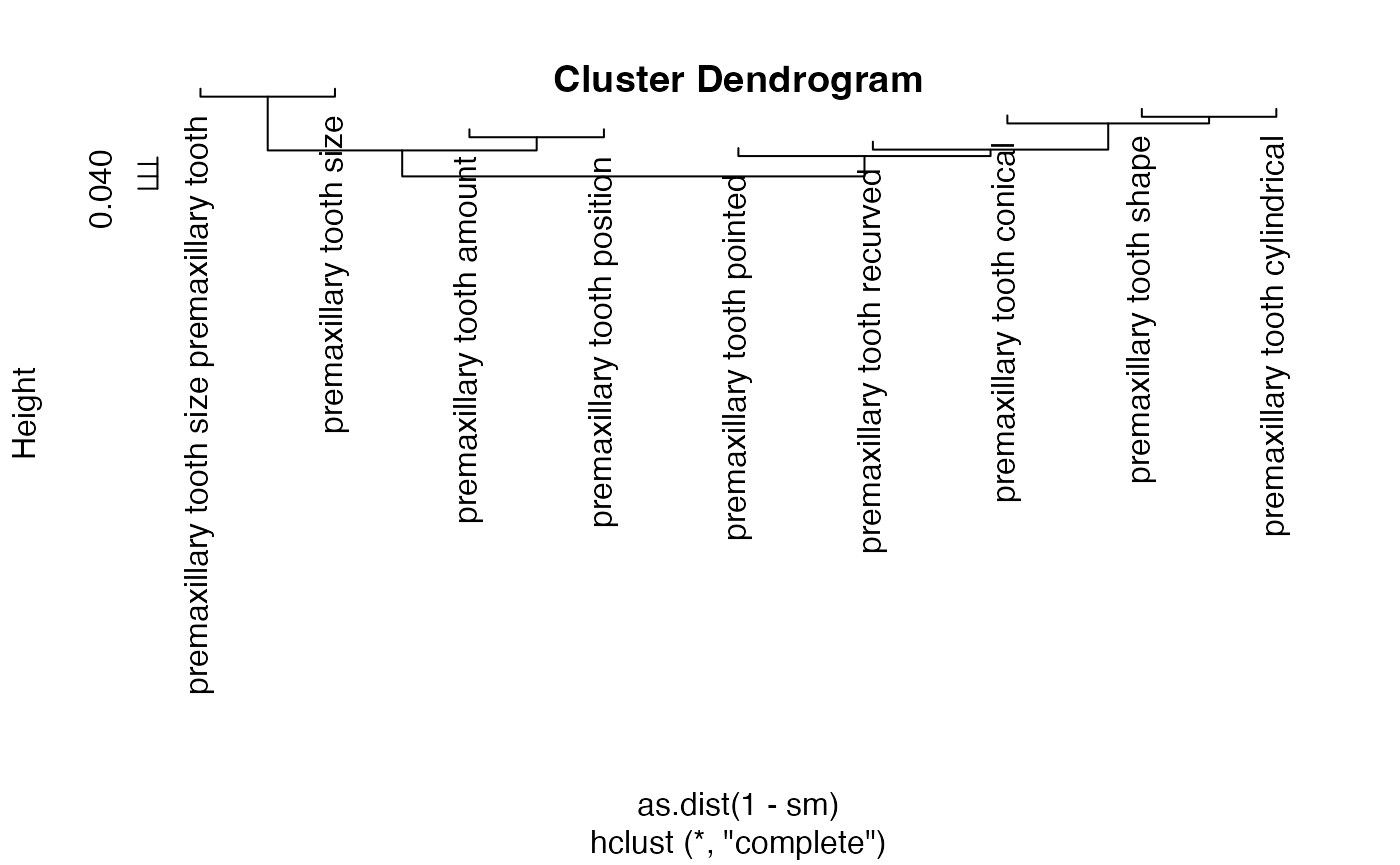
In the PARAMO tutorial, we’ll use a semantic similarity matrix to visualize traits in a heat map. We are also developing methods that use semantic similarity to cluster phenotypes into characters and character states.